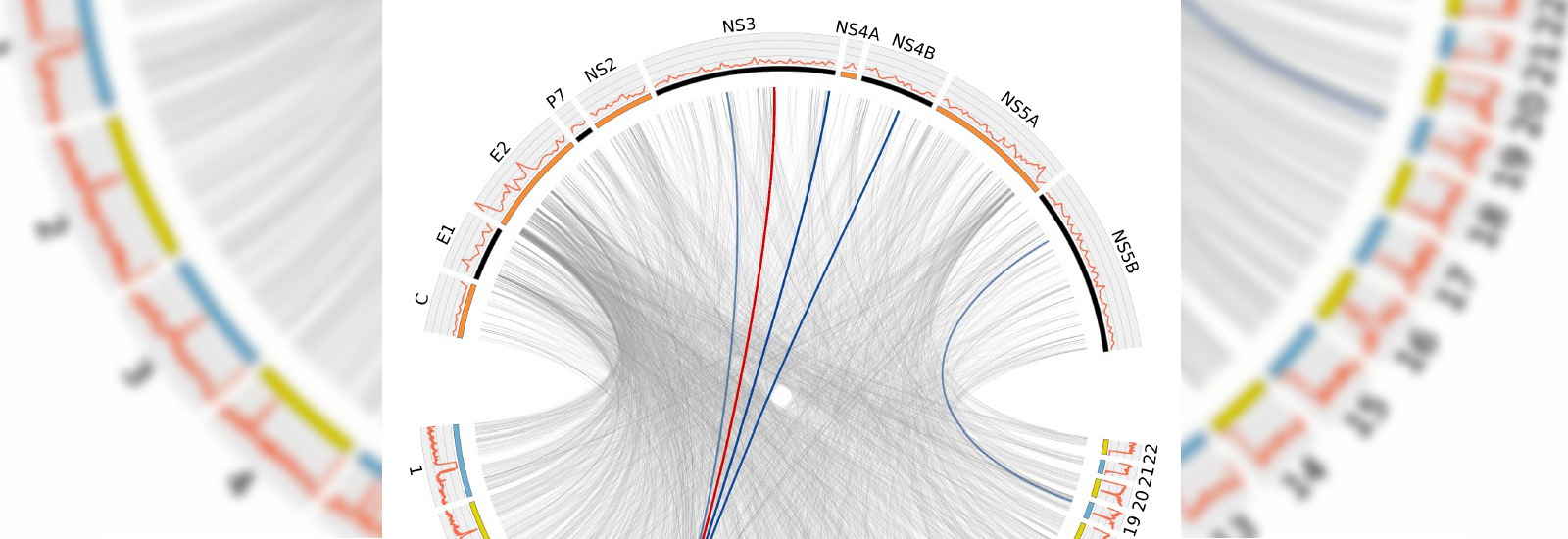
A big data study of hepatitis C and more than 500 patients with the virus has opened the way for a better understanding of how the virus interacts with its human hosts.
Researchers at the University of Oxford have for the first time developed a method for analysing and comparing the genetic makeup of the hepatitis C virus (HCV), as well as that of more than 500 patients with the virus. Looked at together, this will give researchers new insights into HCV and how the human genome interacts with and changes the virus.
Viral hepatitis is one of the leading causes of death and disability worldwide, with 2-3% of the world’s population thought to be infected with HCV, including an estimated 300,000 in the UK. Many people are unaware they are infected with the virus, which left untreated can led to liver disease and cancer.
Professor Ellie Barnes from the Nuffield Department of Medicine, who led the study with Dr Chris Spencer, said: 'This is the first use of a big-data study to look at a virus and host together. We identified two places in the human genome where the genetic variation that calibrates our immune system affects the genetic diversity of the virus.
'There are new drugs available which can clear HCV infection, but they are very expensive and access to them is currently limited. These drugs are also less effective in treating some of the seven strains of the infection that exist than others, which highlights the importance of understanding the genetic basis of the disease for developing future treatments.
'Within 15 years, DNA sequencing of disease-causing bugs like HCV will become a routine part of healthcare. This sort of information can be used to tailor treatments for each patient, to help ensure that the patient is given the best drugs or the right amount of time to give the best possible chance of managing or recovering from infection.'
Researchers within STOP-HCV, a national consortium funded by the Medical Research Council (MRC) and led from Oxford University, are developing big genetic data sets to improve treatments for HCV, and to better understand the biology of HCV infection.
The study highlighted two human genes that change the hepatitis C virus over time.
One of the positions in the human genome (the HLA) reveals those parts of the HCV virus that have tried to mutate to escape recognition by the human immune system. The researchers used the data to create a map of these selective pressures across the viral genome, which can be used to find weak spots that are critical for the virus to survive and these can be used as the target of new therapies or vaccines.
The second position in the human genome highlighted in the study that impacts on the virus activates an immune gene which is switched off in some individuals. The impact of activating this gene is to change the number of viruses that are present in the patient’s blood.
Large-scale analysis of the DNA of the patient and the virus will provide an opportunity to study the way the human immune system naturally responds to HCV infection, and how that impacts on viral evolution.
The full paper, ‘Genome-to-genome analysis highlights the impact of the human innate and adaptive immune systems on the hepatitis C virus,’ can be read in the journal Nature Genetics.